An R package for divergence analysis of omics data
An R package for divergence analysis of omics data
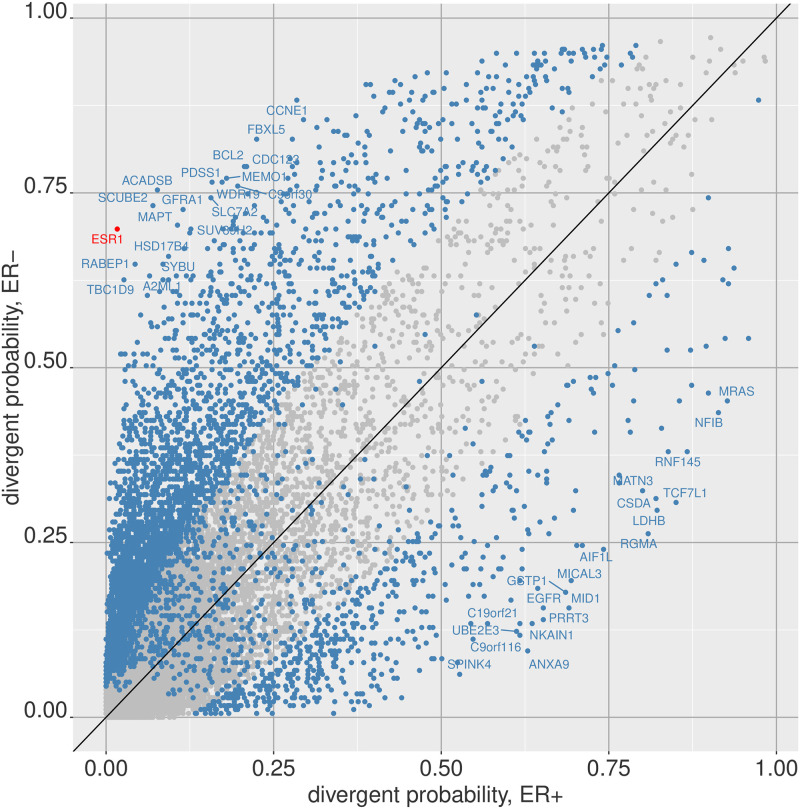
Given the ever-increasing amount of high-dimensional and complex omics data becoming available, it is increasingly important to discover simple but effective methods of analysis. Divergence analysis transforms each entry of a high-dimensional omics profile into a digitized (binary or ternary) code based on the deviation of the entry from a given baseline population. This is a novel framework that is significantly different from existing omics data analysis methods: it allows digitization of continuous omics data at the univariate or multivariate level, facilitates sample level analysis, and is applicable on many different omics platforms. The divergence package, available on the R platform through the Bioconductor repository collection, provides easy-to-use functions for carrying out this transformation. Here we demonstrate how to use the package with data from the Cancer Genome Atlas.